New frontiers in the allele-specific RNA editing research
The regulation of biallelic gene expression is important for development in diploid organisms and is associated with several human diseases. A large class of genes has been demonstrated to be highly expressed in only one allele including random monoallelic expression, parental-specific (imprinted) expression, and sequence allele-specific expression.
Recent studies indicated that allele-specific DNA methylation is one of the factors that influence allele-specific expression. However, such studies mainly focused on the DNA level. As we known, RNA plays the link role in the process of transcription and translation, and various RNA modifications have been reported. However, whether modifications occurring at the RNA level harbor allele-specificity across tissues and species is currently unknown.
RNA editing is a common modification at RNA level that has important implications on the organism’s genetic diversity. The team led by Prof. ZHANG Yaping from Kunming Institute of Zoology, Chinese Academy of Sciences, studied allele-specific RNA editing in various tissues of human and mouse.
They documented 315 allele-specific RNA editing sites, a large proportion of which overlap between transcriptomes from different human tissues. Meanwhile, 184 allele-specific editing sites were identified in brain tissues of heterozygous mice generated by crosses between divergent mouse strains, and a large proportion of these sites overlap in different samples.
Their findings suggest that their method of identifying allele-specific RNA editing is credible, and also indicate that allele-specific RNA editing is ubiquitous. Interestingly, some allele-specific RNA editing sites caused amino acids changes, whereas, a SNP (Single Nucleotide Polymorphisms) adjacent to one of these sites bears synonymous mutational effects.
They conducted cellular experiments which revealed that the synonymous mutation affects the nonsynonymous editing efficiency (Figure 1). This highlighted the likelihood of the synonymous SNP leading to amino acid changes through allele-specific RNA editing.
Furthermore, SHAPE experiment indicated that allele-specific RNA editing was influenced by differences in local RNA secondary structure generated by SNPs. The study represents an exemplary allele-specific RNA editing research and provides a new understanding about connection between synonymous mutations and both diseases and complex traits.
This work was published in Nucleic Acids Research (https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gky613/5052368). Dr. ZHOU Zhongyin (Assistant Investigator at KIZ), Dr. HU Yue (Qilu University of Technology), Dr. LI Aimin (Xi’an University of Technology), LI Yingju (Master’s student at Yunnan University) and Prof. ZHAO Hui (Yunnan University) are co-first authors of this work. Prof. ZHANG Yaping is the corresponding author. This work was supported by grants from Ministry of Agriculture of China and National Natural Science Foundation of China.
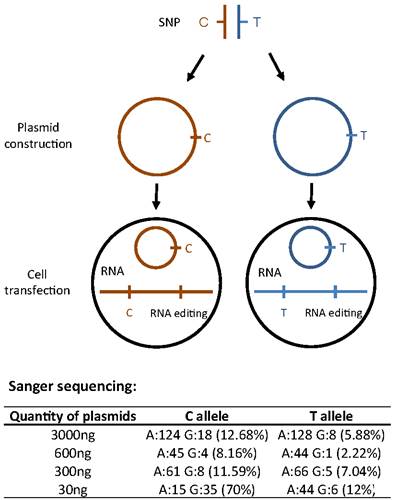
Figure 1. Allele-specific RNA editing causes an amino acid change in the product of the Dact3 gene in the mouse (Image from ZHANG Yaping’s team)
By ZHOU Zhongyin, Editor: HE Linxi
Contact:
HE Linxi
helinxi@mail.kiz.ac.cn
