A collaborative research team led by Prof. SU Bing from Kunming Institute of Zoology, Chinese Academy of Sciences, Prof. LI Cheng from Peking University, and Prof. ZHANG Shihua from Academy of Mathematics and Systems Science, Chinese Academy of Sciences, for the first time, reported by far the highest resolution 3D genome of primate brain, and through cross-species multi-omics analyses and experimental validations, they demonstrate the molecular regulatory mechanisms of human brain evolution. The finding was published in Cell on January 27, 2021.
The unique pattern of human brain development stems from the accumulated genetic changes during human evolution. Among a huge number of diverged genetic changes, only a small portion of the between-species changes are functionally important. The challenge is to identify the causal changes responsible for the unique pattern of human brain development and their regulatory mechanisms. Macaque monkeys, genetically similar to humans, are the ideal model to study the origin and developmental mechanisms of the human brain.

Fig.1 The picture shows a meditating macaque, symbolizing the change of brain and mind during primate evolution. (Image by LUO et al)
Fortunately, as the close relatives of humans, the nonhuman primates (e.g. macaque monkeys) can serve as valuable animal models for probing this task. Macaque monkeys are genetically similar with humans, and they are the ideal model in studying the origin and developmental mechanisms of the human brain.
The genome of mammalian species including human is about 2 meters long and it is compiled in the nucleus with a diameter of only 10 micrometers. The compilation of the genome in the nucleus is not random with organized three-dimensional (3D) distribution, which is important for cell proliferation and differentiation during development. Recently, the invention of whole-genome chromosomal structure capture technology (referred to as Hi-C) provides great opportunity to dissecting the fine-tuned organization of the genome during brain development.
In the published study, researchers conducted cross-species analyses of brain 3D genomes through cross-disciplinary collaboration. With the use of the Hi-C technique, they first constructed a high-resolution 3D chromatin structure map of the macaque fetal brain. Reaching a 1.5kb resolution, this Hi-C map is the highest resolution of primate brains, a highly useful omics dataset for revealing the 3D genome organization of primate brains in details.
At the same time, they also generated the transcriptome map, the chromatin open region map and the map of the anchor protein CTCF. By combining these multi-omics data, for the first time, hey constructed a fine map of chromatin structure of the macaque fetal brain and they identified chromatin structure in different scales, including compartments, topologically associating domains (TADs) and chromatin loops, as well as the regulatory elements in the genome such as enhancers.
In combination with the published brain Hi-C data of human and mouse, they performed cross-species comparison, and they discovered many human-specific chromatin structural changes, including 499 human-specific TADs and 1266 human-specific loops.
Notably, they find that the human-specific loops are enriched with enhancer-enhancer interactions, representing the origin of a fine-tuning mechanism of brain development during human evolution.
Importantly, based on the analysis of single-cell transcriptome data of human brain development, they observed that these human-specific-loop related genes are highly expressed in the subplate lamina, a transient zone of the developing brain critical for neural circuit formation and plasticity.
In the developing human brain, the subplate lamina shows an extradentary expansion compared to macaque and mouse, which is about four times thick of the cortical plate. The subplate starts to decrease after birth and eventually disappears, and we know little about this transient zone. Hence, this study provides the first evidence for the key role of subplate in forming the human-specific brain structures during development.
In addition, the researchers discovered many human-specific mutations (e.g. point mutations and structural changes) are located in the TAD boundary and loop anchor regions, which may lead to the origin of novel binding sites of transcriptional factors and the human-specific chromatin structures. They studied an example involving the EPHA7 gene, which is highly expressed in the subplate and critical for neuronal dendrite development. There are multiple human-specific point mutations in the upstream of EPHA7, leading to the formation of the human-specific enhancers and loops. Through the experiment of enhancer knockout in cell lines, they proved that the human-specific EPHA7 enhancers can cause regulatory changes of EPHA7 expression and affect dendrite development.
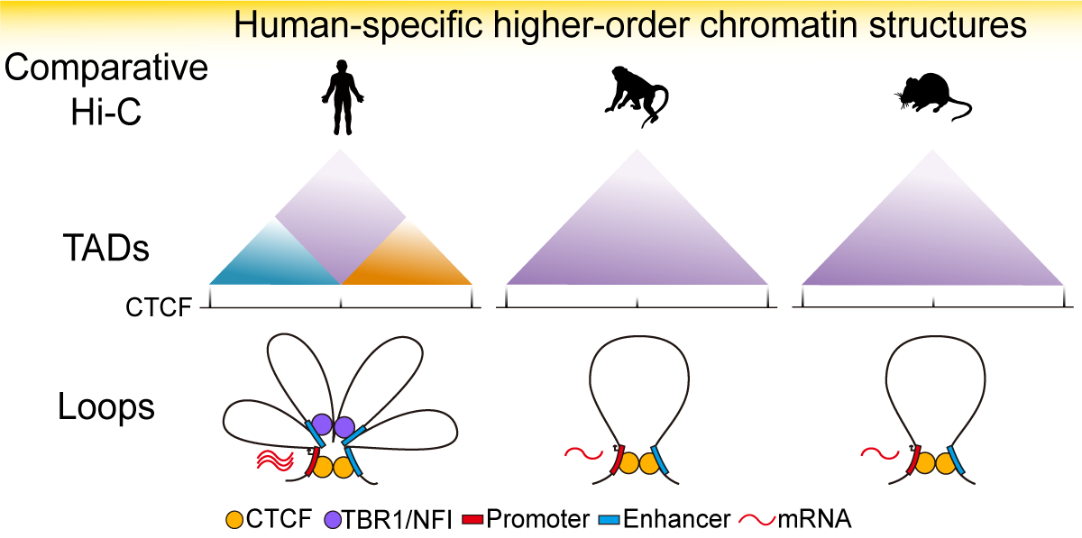
Fig.2 By comparing human, rhesus monkey and mouse, the human-specific higher-order chromatin structures are revealed, which lead to human-specific gene regulatory changes during brain development. (Image by Luo et al.)
This study sheds new light on the genetic mechanism of human brain origin and serves as a valuable resource for brain 3D genomes. The article, entitled “3D Genome of macaque fetal brain reveals evolutionary innovations during primate corticogenesis”, was published on Cell, and the weblink is https://www.cell.com/cell/fulltext/S0092-8674(21)00001-5. Dr. SU Bing, Dr. LI Cheng and Dr. ZHANG Shihua are the co-corresponding authors. This study was financially supported by research grants from Chinese Academy of Science, Ministry of Science and Technology, National Natural Science Foundation of China and Yunnan Provincial funding agencies.
(By SU Bing, Editor: YANG Yingrun)
Contact:
YANG Yingrun
yangyingrun@mail.kiz.ac.cn
