Dr. Peng Shi, Principal Investigator. There are two major topics in our lab. (1) Exploring the genetic mechanisms of animals adaptation to the extreme environments using large-scale data. (2) Functionally resolving the molecular mechanisms of adaptive evolution for animals using functional platforms. My lab investigates these questions using a combination of empirical and computational approaches to analyze the large-scale data.
Email: ship@mail.kiz.ac.cn
Large-Scale Survey of Gut Microbiota Associated With MHE Via 16S rRNA-Based Pyrosequencing
By using 16S rRNA-based pyrosequencing and multiple comparative analyses, we found the continuous overrepresentation of two bacterial families, Streptococcaceae and Veillonellaceae, in cirrhotic patients with and without MHE, compared with normal individuals. In addition, we also discovered an MHE-unique interplay pattern of gut microbiota largely influenced by the members of those two families. Following these findings, we further revealed that gut urease-containing bacteria Streptococcus salivarius was absent in the normal group but was present in cirrhotic patients with and without MHE. The abundance of S. salivarius was significantly higher in cirrhotic patients with MHE than in those without (P = 0.030), and the change in the amount of this bacteria was positively correlated with ammonia accumulation ( R = 0.58, P = 0.003) in cirrhotic patients with MHE but not in those without. Gut microbiota dysbiosis may be associated with the presence of MHE in cirrhotic patients, in particular with ammonia-increasing phenotype in MHE. Gut ammonia-increasing bacteria S. salivarius might be expected to be a potential biomarker of ammonia-lowering therapies in cirrhotic patients with MHE.
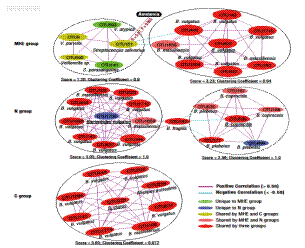
Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study
Elucidating gut microbiota in gallstone patients and complex bacterial colonization of cholesterol gallstones may help in predicting and lowering the risk of cholelithiasis. We describe the composition of bacterial communities of gut, bile, and gallstones in 29 gallstone patients and of gut in 38 normal individuals, based upon large-scale 454 sequencing of bacterial 16S rRNA genes of all 120 samples. Conclusively, this study for the first time discovered gut microbiota dysbiosis potentially associated with cholesterol gallstones. This study further showed a first large-scale glimpse of biliary tract microbiota associated with cholesterol gallstones, ultimately which provides potentially biological and medical implications about the role of bacteria in the formation cholesterol gallstones.
Staff
Dr. Zhen Liu, Associate Professor
Dr. Zhi-Gang Zhang, Associate Professor
Dr. Hui Yang, Assistant Professor
Dr. Jun-Jun Hao, Assistant Professor
Dr. Zhi-Qiang Ye, Assistant Professor
Dr. Ling-Ling Hu, Assistant Professor
Rui Yu, Assistant Experimentalist
Hai-Qing Yu, Assistant Experimentalist
Xing Zhou, Assistant Experimentalist
Graduate Students:Wei Wang, 2009;Dong-Ming Xu, 2010;Fei-Yan Qi, 2011;Jie Luo, 2011; Zhi-Zhong Zheng, 2012;Song-Lin Yang, 2012; Yun Li, 2013;Yu-Long Guo, 2013
