Recently,by using a systematic integrative analysis based on the largest collection of brain expression profiles of Alzheimer’s disease (AD) and other AD related omics data, researchers identified several upstream regulator genes which may play a crucial role in regulatory network of gene expression in AD brain. The study suggested that targeting the expression of these upstream regulators in early stage of the disease may reverse the disease progression.
The research group led byProf. YAO Yonggang at Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS) has published this new study in the journalAlzheimer’s & Dementia(http://www.alzheimersanddementia.com/article/S1552-5260(17)33707-X/fulltext).
AD is the most prevalent neurodegenerative disease in the elderly, and is featured by abnormal aggregation of plaques and tangles in brain, progressive memory loss and cognitive impairment.
Age is the most important risk factor for AD. The incidence increases exponentially with age and doubles every 5 years after age 65. With the progress of global aging, the number of AD patients increases dramatically around the world, which causes heavy burden on families and society.
Besides age, genetic factor is another crucial risk factor. Despite remarkable advances have been made in the understanding of the genetic basis of AD thanks to previous efforts in genetic studies of AD, the pathophysiology of AD is not well understood.
It is reported that over 80% of genetic risk loci identified by genome wide association studies (GWASs) are located in non-coding regions of the genome. These loci are likely to contribute to AD by altering gene expression.
Therefore, a complete characterization of the transcriptomic alterations and the regulatory mechanisms that underpin AD may provide essential evidence to fill the gap between genetic variations and AD development.
In this study, researchers including XU Min and Dr. ZHANG Deng-Feng, together with their colleagues inProf. YAO Yongang’s Group,performed an integrative analysis of available high-throughput brain expression profiling datasets from AD patients and controls using a convergent functional genomic method.
They provided a complete and robust list of differential expressed genes (DEGs) in AD brain, and identified several candidate upstream regulators in the gene expression network, such asYAP1.
Functional experiments ofYAP1have suggested that expression perturbations of upstream regulator genes at the early stage of AD development might be damaging, by affecting expression of downstream genes and so further promoting AD progression. Candidate upstream regulators found in this study might act as potential therapeutic targets for the treatment of AD.
To promote public sharing of AD-related data, they constructed a webserver AlzData (http://www.AlzData.org) for easy access to their results and other high throughput data.
This study was supported by the Key Program of National Natural Science Foundation of China (NSFC, 31730037), the Bureau of Frontier Sciences and Education (QYZDJ-SSW-SMC005), the Strategic Priority Research Program (B) (XDB02020003), and the West Light Foundation of the Chinese Academy of Sciences.
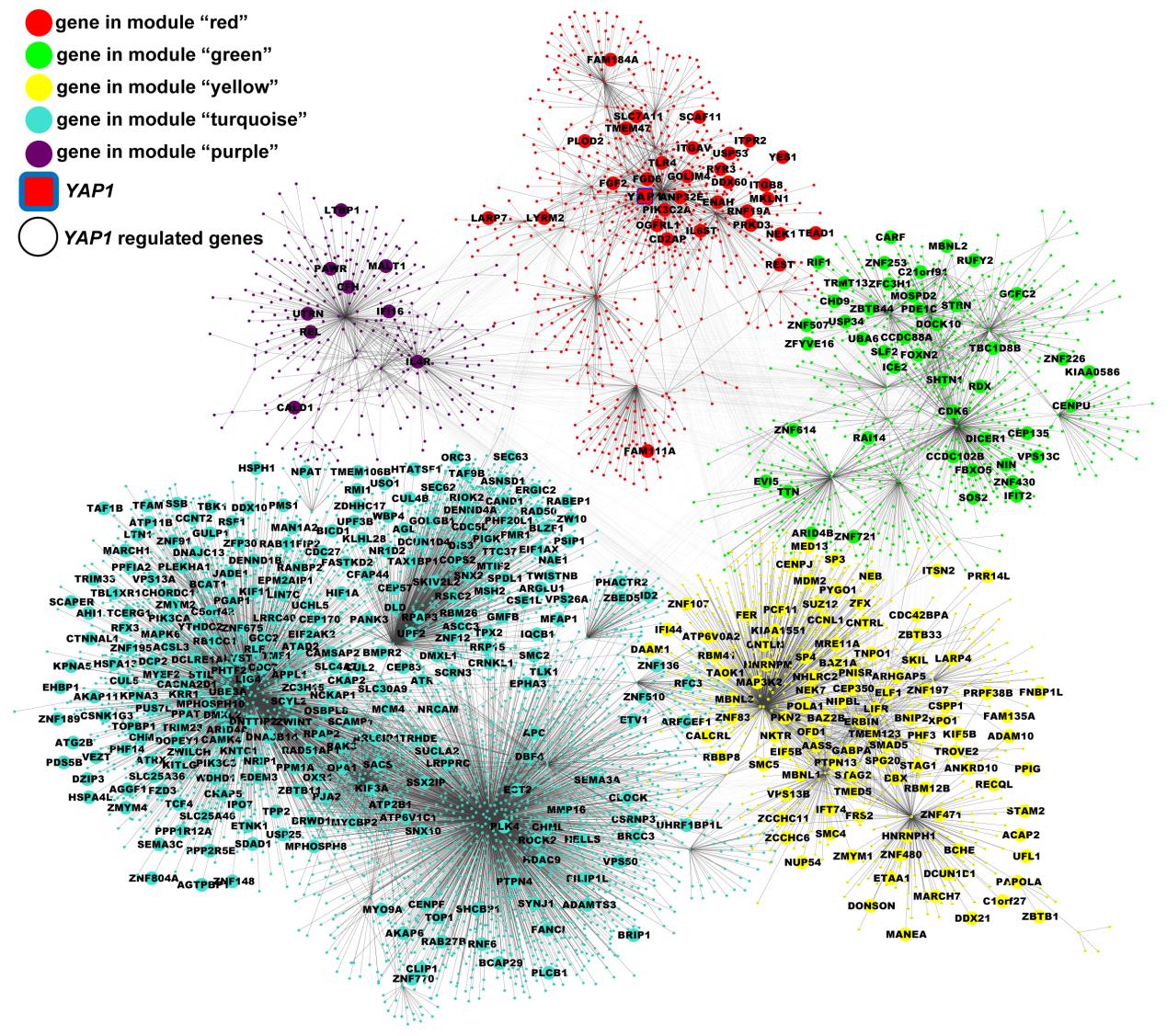
Fig. 1. Expression perturbations ofYAP1could influence the whole AD transcriptional network (Image by YAO Yonggang’s group)

Fig .2. Homepage of AlzData (Image by YAO Yonggang’s group)
(By XU Min, Editor: HE Linxi)
Contact:
HE Linxi
helinxi@mail.kiz.ac.cn

