Heterogeneity is a basic characteristic of cancer, including intra-tumor heterogeneity and heterogeneity among patients mainly due to a variety of genetic alterations. The latter may give rise to differences in the responses to therapies. Glioblastoma (GBM) is the most common and malignant tumor of the brain parenchyma. The median survival of GBM patients is only 12–15 months, although the prognosis of many cancers has significantly improved over the last several decades. Researchers have found that high heterogeneity of glioblastoma contributes to the poor and variant prognosis. However, the existing classification method according to histopathological feature is not enough to solve the problems related to heterogeneity. A team of scientists from the Kunming Institute of Zoology of the Chinese Academy of Sciences (KIZ, CAS) and Columbia University recently found a new method for GBM based on the genetic alterations of CDKN2A and TP53 genes with mutation rates of 60% and 30% respectively. The patients’ survival and gene expression pattern show great difference between two groups. Additionally, the activation of RAS pathway plus Cdkn2a/b silencing can induce GBM, in a similar way to tumor induction by RAS activation plus Tp53 silencing. The corresponding mouse glioblastoma model shows similar difference in expression pattern like humans, which provide strong evidence for the key role of CDKN2A and TP53 in subtyping. This work was published in Oncogene (https://www.nature.com/articles/s41388-018-0305-1). Dr. ZHOU Xia (doctor’s student at KIZ), Dr. LI Gonghua (Associate Investigator at KIZ) and AN Sanqi (Master’s student at KIZ) are co-first authors of this work. Dr. ZHAO Xudong (Principal Investigator at KIZ), Dr. HUANG Jingfei (Investigator at KIZ) and Dr. Antonio Iavarone (professor at Columbia University) are corresponding authors. This work was supported by the National Natural Science Foundation of China, the Top Talents Program of Yunnan Province China, Yunnan Applied Basic Research Projects. 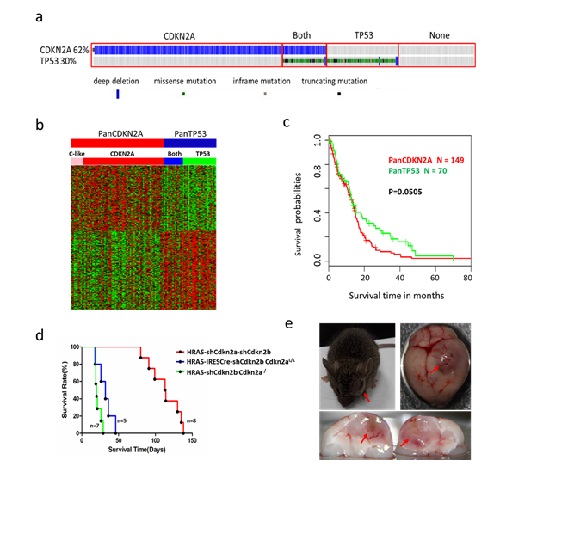 (By ZHOU Xia; Editor: HE Linxi) Contact: HE Linxi helinxi@mail.kiz.ac.cn |

