Schizophrenia is a complex and heterogeneous disorder with a heritability about 80%. In recent years, benefiting from the results of large-scale research consortia and the development of sequencing technology, the genetics study of schizophrenia has made great progress. Genetic studies, including genome-wide association studies (GWASs), whole exome/genome sequencing (WES/WGS), and copy-number variants (CNVs) studies, have identified hundreds of risk genes associated with schizophrenia. However, how to efficiently use these published data to aid schizophrenia genetic study remains a major challenge.
By integrating multiple types of data from various layers of schizophrenia research, scientists from Kunming Institute of Zoology, Chinese Academy of Sciences (KIZ, CAS) developed a schizophrenia genetic database named “SZDB” (www.szdb.org/SZDB) (Schizophrenia Bulletin, 2017) previously. However, there are several limitations of SZDB. First, the gene scoring algorithm arbitrarily treated all the evidences equally. Second, all the gene expression studies were based on microarray technology and the sample size is small. Recent large-scale gene expression studies which based on high throughput sequencing, were not collated in the SZDB. Third, large-scale genetics studies, like GWASs, CNV studies, WGS/WES studies, have identified many novel loci or genes related to schizophrenia in recent years after the development of SZDB. For these many reasons, there is a pressing need to update the SZDB.
Recently, the reseachers released an updated version of a comprehensive database for schizophrenia research, SZDB2.0 (www.szdb.org) with significant data expansion and feature improvements, as well as functionality optimization. Compared with the first version (SZDB), the current database has the following updates: (1) collected the newly published largest GWAS results of schizophrenia; (2) replaced the microarray gene expression data with high throughput sequencing data; (3) included more WGS/WES data of schizophrenia; (4) integrated two polygenic risk score calculators; (5) refined the gene scoring system; (6) collated more CNV publications about schizophrenia; (7) updated other data, including gene expression quantitative trait loci (eQTL), transcript QTL, methylation QTL, and protein–protein interaction data. In all, the current version of SZDB2.0 included most of the large-scale genetics data of schizophrenia which published in the recent years. Moreover, they also optimized the query interface of SZDB2.0 for a better visualization and data retrieval. The updated SZDB2.0 will advance the research of schizophrenia.
This work was published inHuman Geneticsjournal with the title of “SZDB2.0: an updated comprehensive resource for schizophrenia research” (https://link.springer.com/article/10.1007/s00439-020-02171-1). Dr. WU Yong (KIZ, CAS) is the first author of this work. Prof. LUO Xiongjian (KIZ, CAS) and Prof. YAO Yonggang (KIZ, CAS) are the corresponding authors. This study was supported by the Strategic Priority Research Program (B) of the Chinese Academy of Sciences, the National Natural Science Foundation of China, and the Bureau of Frontier Sciences and Education, Chinese Academy of Sciences.
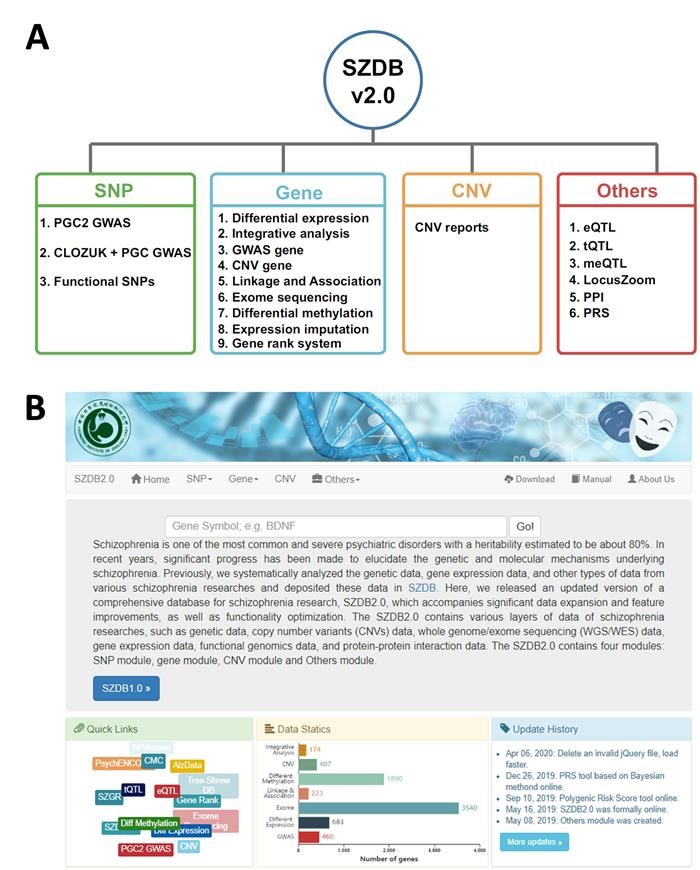
Overview of the updated schizophrenia research database SZDB2.0 (www.szdb.org). a Database structure. b Homepage. (Image by WU Yong)
(By WU Yong; Editor: HE Linxi)
Contact:
HE Linxi
helinxi@mail.kiz.ac.cn

