No matter you like it or not, natural selection is already the key word in modern biology and you can find the signature of it at almost every corner of life science. Mitochondria are the powerhouse of eukaryotic cells. mtDNA is the structure converts the chemical energy into a form cells can use, ATP. Then, how does natural selection manipulate mitochondrial DNA (mtDNA) in tumorigenesis? Dr. KONG Qing-Peng (Kunming Institute of Zoology, the CAS) and his colleagues’ research could give you some clues.
According Dr.KONG’s recent research, the observed mutational pattern on the cancerous mtDNAs might be best explained as relaxation of negative selection.But, in Dr. ZHIDVOK’s study, 2009, the mutation patterns of mtDNAs supposed to be right of the opposite. A significant portion of the cancer mutations was organized in recurrent combinations (COMs), reaching a length of seven mutations, which also colocalized with ancient variants. Thus, by analyzing similarities rather than differences in patterns of mtDNA mutations in tumor and human evolution, evidence for similar selective constraints could be seen, suggesting a functional potential for these mutations. These conclusions are along with the hypothesis which Nowell popped out in 1976 that the formation of cancer was recognized as a process of somatic evolution driven by natural selection, in which any genetic alterations that confer selective advantage to the fitness of the malignant cell would then be preferentially kept.
To clarify these discrepancies, in this study, a total of 186 entire mitochondrial genomes of cancerous and adjacent normal tissues from 93 esophageal cancer patients were systematically analyzed. As an indicator of selective pressure acting on the protein coding gene, the numbers of synonymous (S) and nonsynonymous (N) substitutions in the mtDNA protein-coding regions from cancerous tissue samples and the general human populations were counted, respectively. The results shows that the N/S ratio in cancerous mitochondrial genomes (1.64) was significantly higher than that in the general populations (0.50; P<0.002), which suggests that the selective pressure on the cancerous mtDNAs is different from that on the mtDNAs from the general human populations.
Meanwhile, additional 1,235 cancerous (nearly) complete mtDNA sequences retrieved from the literature have been taken into account. And the same data sets used by other research groups have been re-considered using the phylogenetic approach. It turns out that only one or none COM has been detected. Based on the solid evidence, Dr.KONG claims that most of the previously observed COMs were in fact attributed to problemsof data quality, for example, sample mix-up or contaminationfrom the exogenous DNA.
In summary, the notion that relaxation of negative selection is responsible in the cancerous mutation of mtDNAs is in good agreement with the observation that aerobic glycolysis, instead of mitochondrial respiration, plays the key role in generating energy in cancer cells. This study not only offers a fresh angle to evaluate the role of mtDNAs in tumorigenesis, but also serves as a paradigm emphasizing the importance of data quality in affecting our understanding of the situation.
The article has been published on Molecular Biology and Evolution (doi:10.1093/molbev/msr290)
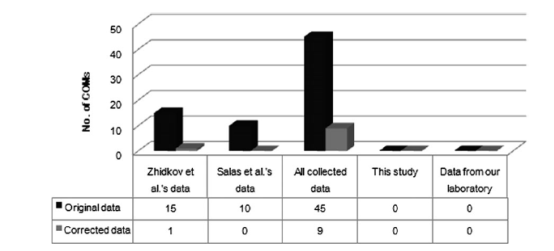
Estimation of COMs based on different cancerous mtDNA data sets. ‘‘Zhidkov et al.’s data’’ refer to the data analyzed in Zhidkov et al. (2009); ‘‘Salas et al.’s data’’ mean the data considered in Salas et al. (2005); ‘‘All collected data’’ indicate all reported cancerous mtDNA considered in the present study (supplementary tables S2 and S3, Supplementary Material online); ‘‘Data from our laboratory’’ represent the cancerous mitochondrial genome data generated by our laboratory, including a total of 123 cancerous mitochondrial genomes from 93 esophageal cancer patients (this study), 10 breast cancer patients (Wang et al. 2007), and 20 colorectal cancer patients (Wang et al. 2011). ‘‘Original’’ means the original data retrieved from the literature, whereas corrected indicates the data corrected by using the phylogenetic approach (figure and legend by KIZ).
