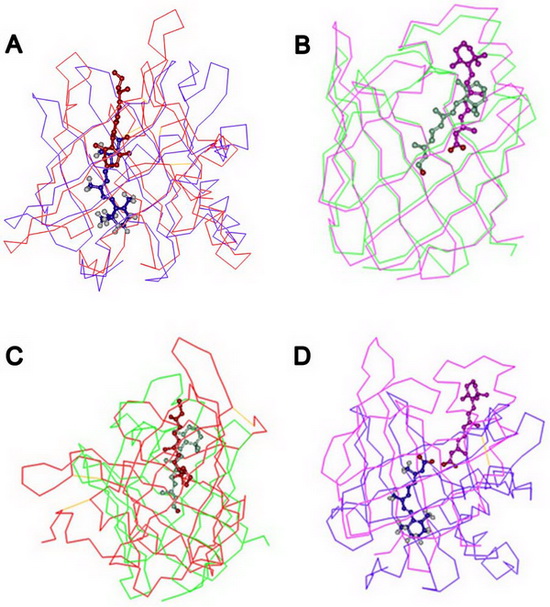
Similar protein architectures can be used to bind identical ligands via completely different ways. Retinoids binding proteins are good case for this principle. Interestingly, although RBP, ERABP, CRBPs and CRABPs have homologous structural motifs and overlapping ligand specificity, they have different binding mechanisms.In the same family of proteins and subcellular location, the orientation of a retinoid molecule withina bindingprotein issame, whereaswhen different families of proteins are considered, the orientation of the bound retinoid is completely different.
In both of the extracellularretinoid-binding proteins (RBP and ERABP), the β-ionone ring of the ligand is positioned in the center of the barrel with the isoprene tail extending along the barrel axis pointing toward the solvent. However, the orientation of the ligand isopposite to that of the corresponding intracellular proteins (CRBPs and CRABPs).In addition, none of the amino acid residues involved in ligand binding is conserved between the transport proteins. However, for each specific binding protein, the amino acids involved in the ligand binding are conserved.
Dr. Yuru Zhang and Yuqi Zhao in professor Huang lab analyzed the various retinoid transport mechanisms through bioinformatic and molecular dynamics simulations methods and propose a possible transport model for retinoids. This study have been published in PLOS onehttp://dx.plos.org/10.1371/journal.pone.0036772。
