When we see the colorful world or smell a pleasant (or repulsive) scent, light or chemical signals are converted into electrical signals in our eyes and noses, signals that our brain recognizes and processes. Crucial for this signal conversion is a group of membrane proteins called cyclic nucleotide-gated (CNG) channels. A team of scientists from the Kunming Institute of Zoology (KIZ) of the Chinese Academy of Sciences (CAS), Columbia University and Tsinghua University recently determined the first high-resolution three-dimensional structure of a eukaryotic CNG channel. This structure provides unprecedented insights into CNG channel properties and a new framework for understanding how CNG channels open and close and how genetic mutations of CNG channels cause blindness. This work was published online in Nature on January 18th.
In vertebrates, vision and olfaction signal transduction relies on CNG channels. In photoreceptors, light activation of the photopigments decreases intracellular cyclic guanosine monophosphate (cGMP) concentration and closes CNG channels, resulting in membrane hyperpolarization. In olfactory sensory neurons, odorant activation of olfactory receptors increases intracellular cyclic adenosine monophosphate (cAMP) concentration and opens CNG channels, leading to membrane depolarization. Mutations in CNG channel genes have been associated with debilitating visual disorders such as retinitis pigmentosa and achromatopsia.
CNG channels are members of the voltage-gated ion channel (VGIC) superfamily. VGICs have two structural and functional modules, a voltage-sensor domain (VSD) or voltage-sensor-like domain (VSLD) that moves in response to membrane voltage changes and a pore domain that opens and closes in response to VSD or VSLD movements. Although CNG channels possess a VSLD, they are not controlled by membrane voltage. This voltage-insensitivity is critical for proper phototransduction and olfactory transduction. Instead, CNG channels are controlled by intracellular cAMP or cGMP, which binds to the cytoplasmic C-terminus.
The KIZ, Columbia and Tsinghua collaborative team used a cutting-edge technique called single particle electron cryo-microscopy (cryo‑EM) to obtain a 3.5 Å‑resolution cGMP-bound open-state structure of a full-length CNG channel from the nematode Caenorhabditis elegans. The structure reveals several novel and/or functionally important features, including a non-domain swapped configuration of its VSLD and pore domain, direct interactions between the C-terminus and both the pore domain and VSLD, and a segmented and restrained S4 transmembrane segment that evidently explains why CNG channels are not controlled by membrane voltage. These and other structural features are in good agreement with CNG channel properties elucidated in extensive previous functional studies and provide new insights into the allosteric mechanisms of how cyclic nucleotide binding opens the channel. The structure also provides a blueprint for understanding and investigating the functional effects of hereditary disease-causing mutations.
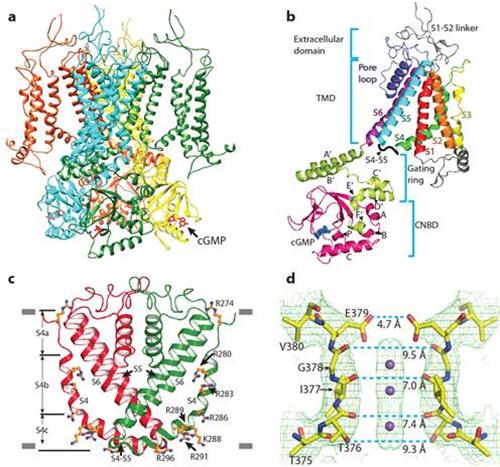
Figure a: structure of a CNG channel tetramer, viewed from parallel to the membrane. Figure b: structure of a CNG channel protomer. Figure c: an unconventional S4 transmembrane segment. Figure d: close-up structure of the ion selectivity filter. In c and d, only two diagonally opposed subunits are shown.
Dr. Minghui Li (Associate Research Scientist in the Department of Biological Sciences of Columbia University and Visiting Scientist of KIZ), Xiaoyuan Zhou (PhD student at Tsinghua University) and Dr. Shu Wang (Associate Investigator in the Ion Channel Research and Drug Development Center (ICDC) of KIZ) are co-first authors of this work. Dr. Jian Yang (Investigator in ICDC and Professor in the Department of Biological Sciences of Columbia University) and Dr. Xueming Li (Professor in the Beijing Advanced Innovation Center for Structural Biology of Tsinghua University) are corresponding authors.
This work was supported by grants to J.Y. from the National Key Basic Research Program of China (2014CB910301), the National Institutes of Health (R01GM085234 and RO1NS053494), the National Natural Science Foundation of China (31370821), the Top Talents Program of Yunnan Province (2011HA012) and the High-level Overseas Talents of Yunnan Province; to X.L. from the China Youth 1000-Talent Program of the State Council of China, Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences and the National Natural Science Foundation of China (31570730); and to S.W. from the Key Research Program of the Chinese Academy of Sciences (KJZD-EW-L03), the National Natural Science Foundation of China (81302865), West Light Foundation of the Chinese Academy of Sciences, Yunnan Applied Basic Research Projects (2013FB074) and the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Link to Nature article:
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature20819.html
(By Jian Yang)
Contact information
Jian YANG
E-mail: jianyang@mail.kiz.ac.cn
