Protective resemblance is an important ecological adaptive mechanism in the struggle for survival of many organisms. It usually includes three categories: general resemblance to the environment (i.e., crypsis), more exactly imitating surrounding objects, such as bark, a leaf, or a flower (i.e., masquerade), and resemblance of other species (mimcry: such as Bayesian mimicry and Mullerian mimicry). Many insects, especially butterflies, exhibit a variety of protective morphological traits, which were used as the important evidence by Darwin and Wallace to support the theory of natural selection. Kallima inachus (Nymphalidae) is one of the textbook species with protective morphology. It is also known as the oakleaf or dead-leaf butterfly because they display transverse, leaf-like venation across the ventral sides of the fore- and hind-wing. Although the dead-leaf butterfly is considered to be a typical representative of the case of masquerade but almost nothing is known about the genetic and molecular basis of its protective morphology.
The research team from Kunming Institute of Zoology, Chinese Academy of Sciences (KIZ, CAS) has long been committed to conduct research on the genetic basic of insect morphological evolution using butterflies as models since 2010. They dissected the genomes of Papilio machaon (type species for all butterflies) and its close species, P. xuthus, for the first time, and successfully realized CRISP/Cas9 gene editing of wild insects using butterfly P. xuthus as a model (Li et al., 2015, Nature Communications). Based on these previous works, on one hand, they continued to combine other multi-omics data and functional experiments to investigate the genetic regulation basis of butterfly morphological evolution. On the other hand, in order to better explore the genetic basis of butterfly diversity at a larger scale of butterfly phylogeny, they started a Butterfly Phylogenome Project in 2017. As the first step of Butterfly Phylogenome Project, they determined the genome C-value of representative butterfly species in various families in China, finding that the genome size of butterfly ancestors is about 500 Megabase (Mb) (Liu et al., Systematic Entomology, 2020). Then, they assembled the first chromosomal-level butterfly (Papilio binaor) genome using Hi-C technology and three-generation long read sequencing technology (Lu et al., GigaScience, 2019).
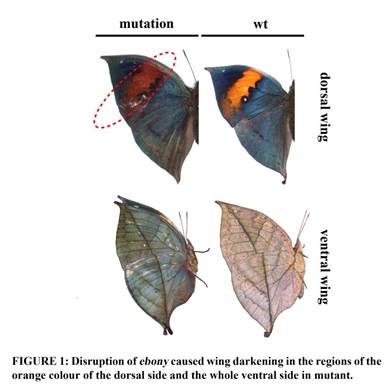
Recently, by combining karyotype experiments, Pacbio sequencing technology and Hi-C technology, they again dissected the genome of K. inachus, a textbook species with protective morphology. They determined the the number of haploid chromosome (n = 31) of the dead-leaf butterfly K. inachus, and generated a high-quality, chromosome-level assembly (568.92 Mb; contig N50: 19.20 Mb). They also identified candidate Z and W sex chromosomes.This is the first butterfly reference genome with with the W chromosome assembled and the most complete butterfly reference genome to date. The assembled genome contains 15,309 protein-coding genes and 49.86% repeat elements. Phylogenetic analysis showed that K. inachus diverged from Melitaea cinxia (no leaf resemblance), both of which are in Nymphalinae, around 40 million years ago. Demographic analysis indicated that the effective population size of K. inachus decreased during the last interglacial period in the Pleistocene. The wings of adults with the pigmentary gene ebony knocked out using CRISPR/Cas9 showed phenotypes in which the orange dorsal region and entire ventral surface darkened, suggesting its vital role in the ecological adaption of dead-leaf butterflies. These results provide important genome resources for investigating the genetic mechanism underlying protective resemblance in dead-leaf butterflies and insights into the molecular basis of protective coloration.
The study, titled Chromosome-level reference genome assembly and gene editing of the dead-leaf butterfly Kallima inachus, was published on Molecular Ecology Resources on May 12, 2020. Website link: https://onlinelibrary.wiley.com/doi/abs/10.1111/1755-0998.13185.
The work was finished by the Insect Research Team from Kunming Institute of Zoology, Chinese Academy of Sciences, led by Dr. LI Xueyan, who is also the first corresponding author of the article. This work was supported by grants from the National Natural Science Foundation of China (No. 31621062), State Key Laboratory of Genetics Resources and Evolution (No. GREKF18-13), “Light of West China” of CAS, Strategic Priority Research Program of CAS (XDB13000000) (to W.Wang), etc.
(Image by Dr. LI Xueyan)
(By LI Xueyan, Editor: HE Linxi)
Contact:
HE Linxi
helinxi@mail.kiz.ac.cn
