Whole-genome duplication (WGD) or polyploidy provides genomic opportunities for evolutionary innovations and adaptation. Polyploid animal appear to be incapable of coping with genomic and developmental chaos resulting from the merging of two genomes in newly synthesized systems. In polyploid plants, biased subgenomic changes may help alleviate chaos from genome mergers, and subsequent co-ordination may help stabilize the subgenomic functions after the initial “genome shock” between divergent subgenomes that coexist in the same cell nucleus. However, consequences of polyploid animals in the long-term rediploidization remain elusive.
Goldfish family (Cyprinidae), the oldest and most common aquarium pet species, includes hundreds of polyploid species. Goldfish has nearly twice as many chromosomes as zebrafish and most of other cyprinids. Its numerous small chromosomes pose a great challenge to assembling and annotating both subgenomes at the chromosomal level. Despite the recently published draft genome, the evolution of the complex polyploid genome remains poorly understood, because no extant diploid progenitors are available in goldfish.
Researchers from Kunming Institute of Zoology and their collaborators recently reported an improved reference-quality de novo genome for allotetraploid goldfish whose origin dates to ~15 million years ago.
Comprehensive analyses identify changes in subgenomic evolution from asymmetrical oscillation in goldfish and common carp to diverse stabilization and balanced gene expression during continuous rediploidization. The homoeologs are co-expressed in most pathways, and their expression dominance shifts temporally during embryogenesis. Homoeolog expression correlates negatively with alternation of DNA methylation.
The results show that allotetraploid cyprinids have a different strategy for balancing subgenomic stabilization and diversification. The diverse and continuous evolutionary processes provide novel insights into genome evolution and function of allopolyploid vertebrates and may explain why most polyploid animals fail to survive.
The study entitled “From asymmetrical to balanced genomic diversification during rediploidization: Subgenomic evolution in allotetraploid fish” been published in Science Advances.
The research work was supported by grants from the National Natural Science Foundation of China, Yunnan Provincial Science and Technology Department, the fund from China Agriculture Research System, Hunan Provincial Natural Science and Technology Major Project, The Second Tibetan Plateau Scientific Expedition and Research Program (STEP), CAS "Light of West China" Program, National Postdoctoral Program for Innovative Talents, and China Postdoctoral Science Foundation.
(Imaged by CHAI Jing and LV Xuemei )
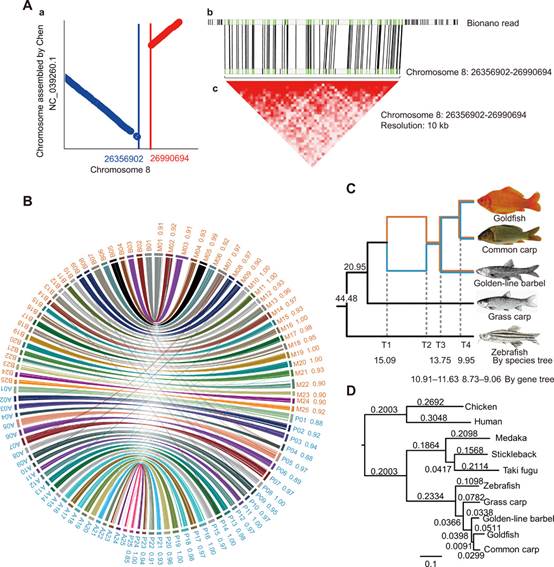
FIGURE 1. Syntenic blocks, phylogeny, subgenome identification, and allotetraploidy. (A) Diagrams displaying the rearrangement identified in the syntenic blocks between our assembly and previously published genome. (B) Allotetraploid goldfish and common carp genome synteny. (C) Phylogenetic relationships and timing of WGD/polyploidization events in Cyprininae, with nodes based on protein-coding genes of goldfish, common carp, golden-line barbel, grass carp, and zebrafish. (D) Phylogenetic tree based on protein-coding genes from single-copy orthologs, rooted with human and chicken. Alignments were performed by MUSCLE, and the maximum likelihood tree was reconstructed by PhyML.
(Image adopted from Fig. 2 of the published paper)
(By CHAI Jing, Editor: YANG Yingrun)
Contact:
YANG Yinrun
yangyingrun@mail.kiz.ac.cn
