| Scientists Reveal Spatial Gene Expression and Regulation in Primate Brains |
| 2018-07-12 | | 【Print】 |
Changes in gene expression were long thought to underlie the rapid evolution of phenotypic traits. While several studies have investigated the expression dynamics across different brain regions as well as the regulatory mechanisms driving human-specific changes in gene expression, no further steps have been taken towards the understanding of human-specific expression across multiple brain regions using the combination of transcriptomics and epigenomics.
In a study published in Genome Research, an international research group led by Prof. Philipp KHAITOVICH from CAS-MPG Partner Institute for Computational Biology of Chinese Academy of Sciences determined uniquely human transcriptome features based on gene expression profiles collected across eight functionally distinct brain regions of humans, chimpanzees, gorillas, and a gibbon.
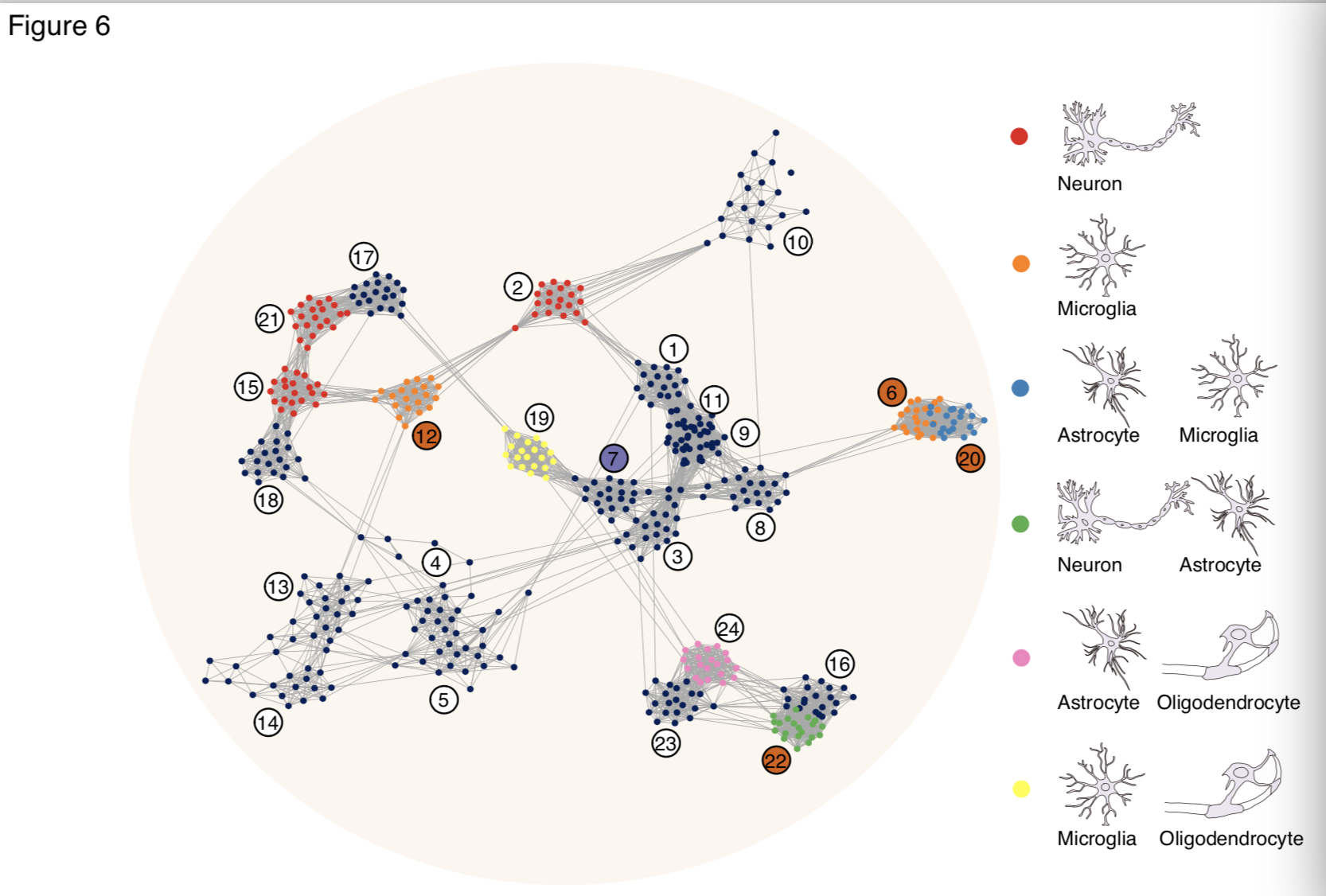
The analysis based on expression profiles across brain regions revealed eight human-specific co-expression modules, four of them supported by both outgroup species.
The four human-specific modules representing expression changes in neocortical areas, cerebellum, and hippocampus contained 1,851 genes cumulatively. These modules contained human-specific changes in the neurons and astrocytes of the human hippocampus, as well as the changes in the microglia of the human neocortical regions and cerebellum. Researchers further assessed and compared regulatory mechanisms that drive gene expression differences among species utilizing the epigenome data collected in eight brain regions of humans, chimpanzees, and macaques.
The results showed that the proportions of gene expression changes assigned to the human and the chimpanzee lineages deviated little from the 50:50 ratio. Furthermore, researchers found that between transcription factors (TFs) and H3K27ac epigenetic modifications, which are associated with active promoters and enhancers, TFs represent the main driving force underlying recent evolutionary changes. By contrast, genes controlled by epigenetic modifications associated with active promoters were the most conserved. This study provided novel insights into the evolution of expression features unique to the human brain transcriptome, as well as potential focal points for further research of the molecular mechanisms underlying the particularity of human brain. |