| Population genomics of wild Chinese rhesus macaques reveals a dynamic demographic history and local adaptation, with implications for biomedical research |
| 2018-12-05 | | 【Print】 |
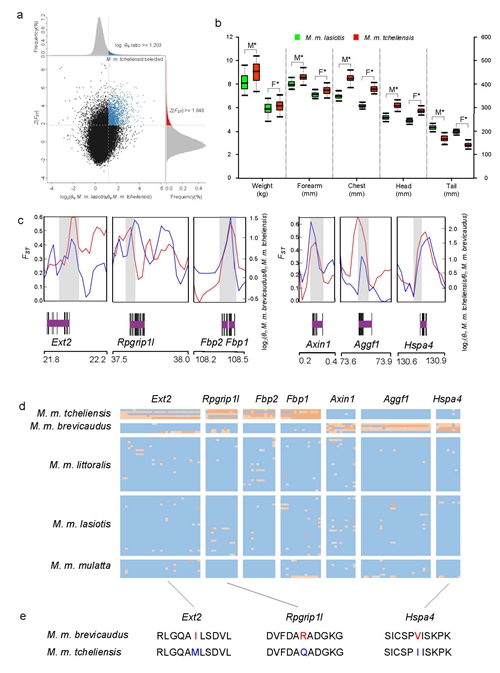
Rhesus macaques (RMs, Macaca mulatta) are the world’s most widely distributed primates, occupying a vast geographic distribution spanning from Afghanistan to the Chinese shore of the Pacific Ocean and south into Myanmar, Thailand, Laos and Vietnam. As the most widely distributed nonhuman primatespecies, RMs occupy diverse ecological landscapes and habitats, making them an interesting model to address questions about how species evolve and adapt to local environmental variation, including characterizing the genomic architecture of adaptation to habitat, climate and medical research. At present, Prof. Li Ming’s team at Institute of Zoology, Chinese Academy of Sciences (IOZ, CAS) and Prof. Michael W. Bruford’ team at Cardiff University, UK, re-sequenced 81 geo-referenced individuals of five subspecies from 17 locations in China using Illumina HiSeq2000 and Illumina HiSeq2500 and conducted the study of its population genomics. The results indicated that the wild RMs populations were structured into five genetic lineages on the mainland and Hainan Island, recapitulating current subspecies designations. These subspecies are estimated to have diverged 109 to 18 thousand years ago, but feature recent gene flow. Consistent with the expectation of a larger body size in colder climates and smaller body size in warmer climates (Bergman’s rule), the northernmost RM lineage (M. m. tcheliensis), possessing the largest body size of all Chinese RMs, and the southernmost lineage (M. m. brevicaudus), with the smallest body size of all Chinese RMs, feature positively selected genes responsible for skeletal development. Further, two candidate selected genes (Fbp1, Fbp2) found in M. m. tcheliensis are involved in gluconeogenesis, potentially maintaining stable blood glucose levels during starvation when food resources are scarce in winter. The tropical subspecies M. m. brevicaudus showed positively selected genes related to cardiovascular function and response to temperature stimuli, potentially involved in tropical adaptation. This study also found 118 SNPs matching human disease-causing variants with 82 being subspecies-specific. This study provides a resource for selection of RMs in biomedical experiments. Also, the demographic history of Chinese RMs, and their history of local adaption in this study offers new insights into their evolution and provides valuable baseline information for biomedical investigation. This work was published in GigaScience (https://academic.oup.com/gigascience/article/7/9/giy106/5079661?searchresult=1). Dr. LIU Zhijin (Associate Professor at IOZ), TAN Xinxin (Doctoral student at IOZ) and Dr. Pablo Orozco-terWengel (Postdoc at Cardiff University, UK) are co-first authors of this work. Dr. LI Ming (Professor at IOZ) and Dr. Michael W. Bruford (Professor at Cardiff University, UK) are corresponding authors. This work was supported by grants National Natural Science Foundation of China (31530068), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000,XDA19050202) and National Key R&D Program of China, 2016YFC0503200l). |