Hair plays an important role in primates and is clearly subject to adaptive selection. While humans have lost most facial hair for better thermal dissipation, eyebrows are a notable exception. Eyebrow thickness is heritable and widely believed to be subject to sexual selection. Nevertheless, few genomic studies have explored its genetic basis.
A research team led by Prof. Sijia Wang from CAS-MPG Partner Institute for Computational Biology at the Chinese Academy of Sciences performed genome-wide association studies on eyebrow thickness in multiple ethnic groups, including Han Chinese, Uyghurs, Latin Americans, and Caucasians. The study reported solid evidence that novel genetic variants near the SOX2, FOXD1 and EDAR genes could affect eyebrow thickness. Researchers prioritized four variants for experimental verification after fine mapping. CRISPR/Cas9-mediated gene editing provided evidence that the variants rs1345417 and rs12651896 affect the transcriptional activity of the nearby SOX2 and FOXD1 genes, respectively. This represents a successful example of a combination of GWAS and CRISPR/Cas9 technology to demonstrate how non-coding variants with regulatory functions may play an important role in common diseases and traits. Finally, statistical analyses suggest that, contrary to popular speculations, eyebrow thickness is not be subject to strong natural selection pressure. This study reveals the genetic mechanism that affects the thickness of human eyebrows, and provides new ideas for further research on hair density.
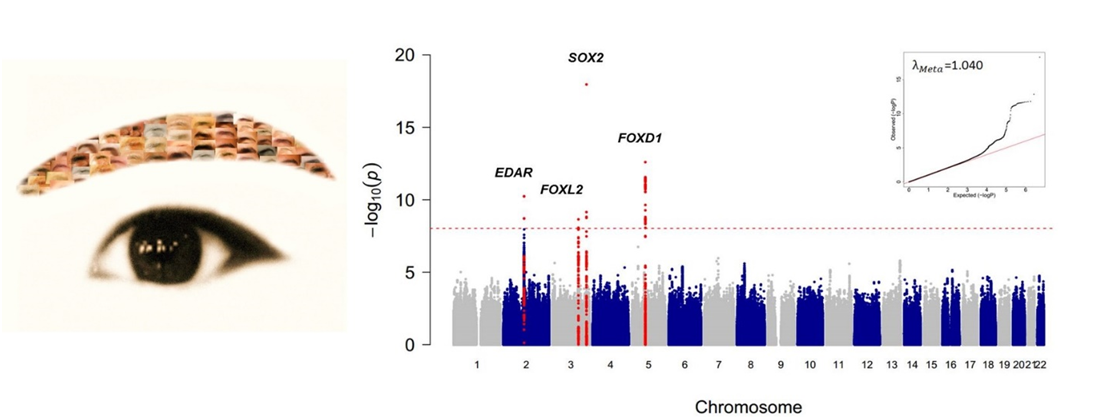
The work was published online in the journal of PLOS Genetic on Sep. 24, 2018, entitled "Genome-wide Association Studies and CRISPR/Cas9-mediated Gene Editing Identify Regulatory Variants Influencing Eyebrow Thickness in Humans'. This study was mainly supported by the "Strategic Priority Research Program" of the Chinese Academy of Sciences, National Natural Science Foundation of China (NSFC), the National Science & Technology Basic Research Project and the Max Planck-CAS Paul Gerson Unna Independent Research Group Leadership Award.